Do you want to know what the Gedmatch One-to-One Autosomal DNA Comparison tool can tell you? In this post, I will discuss what you can learn from using this tool.
There are many reasons that you might want to use the Gedmatch One-to-One Comparison tool. Whether you know you should use it to double-check the results from the One-to-Many tool, or you want to compare your DNA to a specific person, the One-to-One tool provides a wealth of valuable information.

Important: Before you contact any of your DNA matches on the One-to-Many list, you should always run this One-to-One Autosomal DNA Comparison tool first. This way, you will have as much knowledge as possible as to how you might be related. Knowledge is power.
In order to help me explain the results, I have a partial screenshot of a comparison that I ran of my DNA and my dad's first cousin's kit. He is a known first cousin once-removed to me, and he was the first person from my known family to take a DNA test.
Fortunately, my relative is also very interested in learning more about his DNA results, so he has also uploaded his DNA to Gedmatch where we can make more detailed comparisons.
When I first got my Ancestry DNA matches back and this relative showed up in the 1st-2nd cousin category, I knew that this "whole DNA testing thing" was accurate. In this article, I will show you an example of results from the One to One Autosomal DNA tool on Gedmatch and explain what each of the columns mean.
One-to-One Autosomal DNA Comparison tool - the Basics
It is important to first mention that Gedmatch is not a DNA testing company. Instead, you must first take a DNA test with another company. Then, you can download your raw DNA data file and upload it to Gedmatch.
If you have not yet tested your DNA, I would recommend doing so as soon as possible so you can start using Gedmatch. You can read about autosomal DNA testing and how to choose the best DNA test for your needs here:
If you have already tested your DNA with any of the most popular DNA testing companies, you most likely can download your DNA data into a file that can then be uploaded to Gedmatch. All you need to use Gedmatch is a free account that can be created in just a few minutes.
Once you have uploaded your DNA, you can start making comparisons between your DNA and your genetic relatives. The results will show exactly where you and your relative match on each chromosome, as well as details about those shared DNA segments.
The image above is from the comparison between myself and my first cousin. Below, I will jump into the details of understanding exactly what these results tell us.
When I ran the tool for the purposes of writing and updating this article, I selected the "Positions only" option on the tool, instead of graphics and positions. In my opinion, this is easier for beginners to understand, so the results you see below might look different than if you run the tool on the default setting that also shows the visualization of the matching DNA segments.
Numbered chromosome DNA segment data
The most important element of the One-to-One Autosomal DNA Comparison tool results is the detail about the location and size of all shared DNA segments. The default setting for this tool is 7 centimorgans, so the results will show all DNA segments shared between two people that are larger than this default size.
These results are reported in a table format, and can sometimes be accompanied by graphics if you left the tool on the default setting. If you are viewing graphics and position of shared segments, you will likely see multiple tables, especially if you are more closely related to your DNA match.
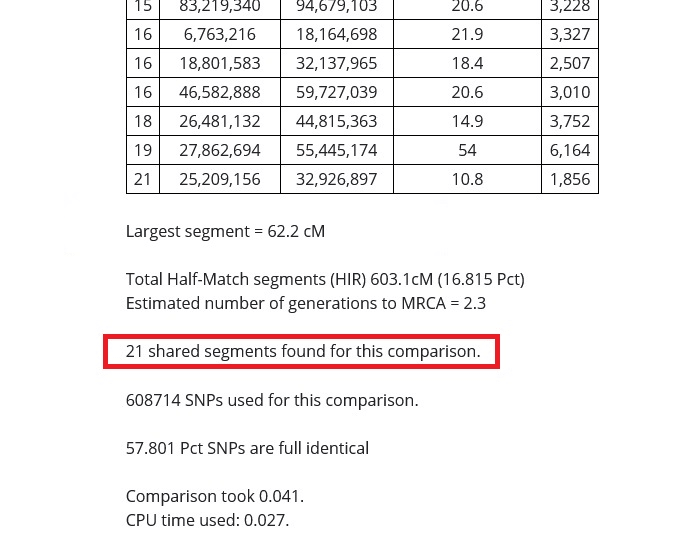
One of the first things that you will notice about your Gedmatch One-to-One Autosomal DNA Comparison tool results is that there will be tables, or multiple tables (if you chose to view the graphics), that have a lot of numbers in them. Some of these numbers are quite large, but it is easy to understand if you know what to look for.
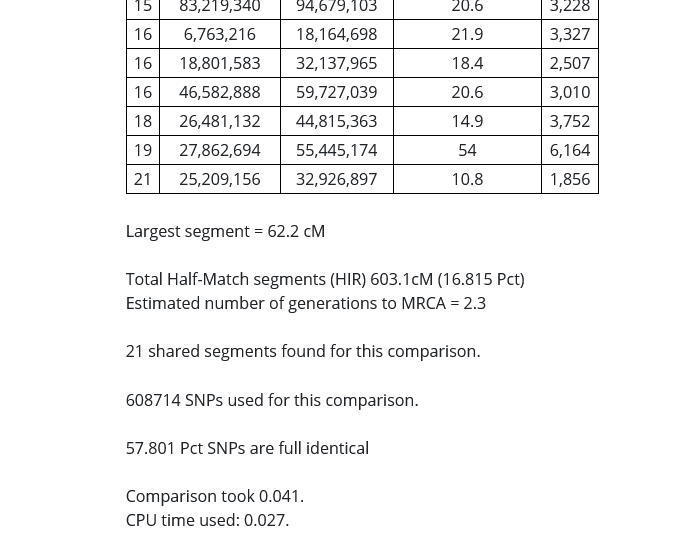
In the image above, I put a red rectangle around the DNA segment that my cousin and I share on Chromosome 19. The table is telling me that my cousin and I share a 54 centimorgan DNA segment on Chromosome 19 that starts at 27,862,694 and ends at 55,445,174, and that 6164 SNPs were examined in the course of the analysis.
Largest segment shared
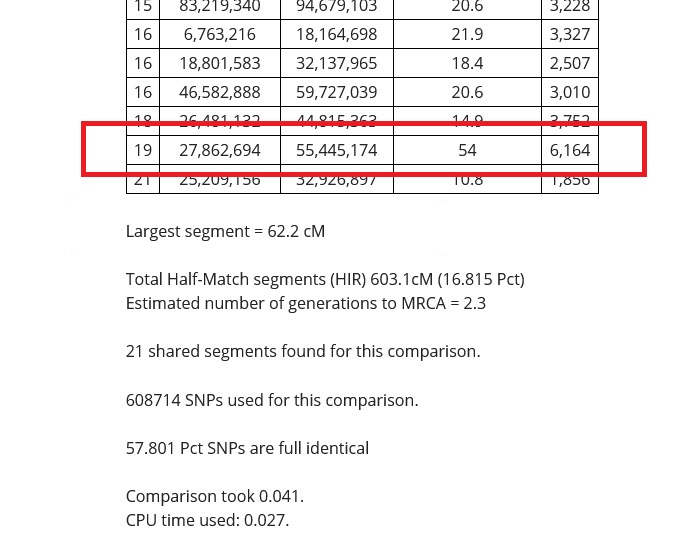
The first line below the table of results shows the size of the largest DNA segment shared. This information is important because it can help us determine how far back we might look in order to find our most recent common ancestor.
In the image below, you can see that my match and I share a DNA segment of 62.2 cMs (centimorgans) in length. This is a large segment, and if you find that you share a segment this large with your match, they are not distantly related to you.
The larger the DNA segment, the closer the relationship. The smaller the segment, the more distant, which means that we might need to look very far back in our family tree to find the common ancestor shared between distant cousins sharing only one or two small segments.
Total half-match segments (HIR)
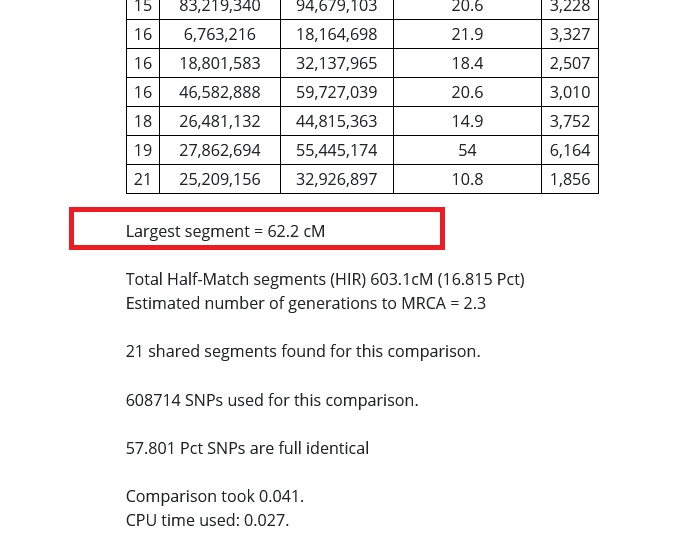
The number that is reported after where it says "Total half-match segments (HIR)" is the total number of centimorgans you and your match share across all of the chromosomes listed in the table. This number is the total amount of DNA that you share with your relative.
In the red box in the image below, you can see that my relative and I share 603.1 cMs (centimorgans) of DNA. If I were to get a calculator and add up the centimorgans shared on every segment on every chromosome reported in the tables on the results, this number should match almost exactly.
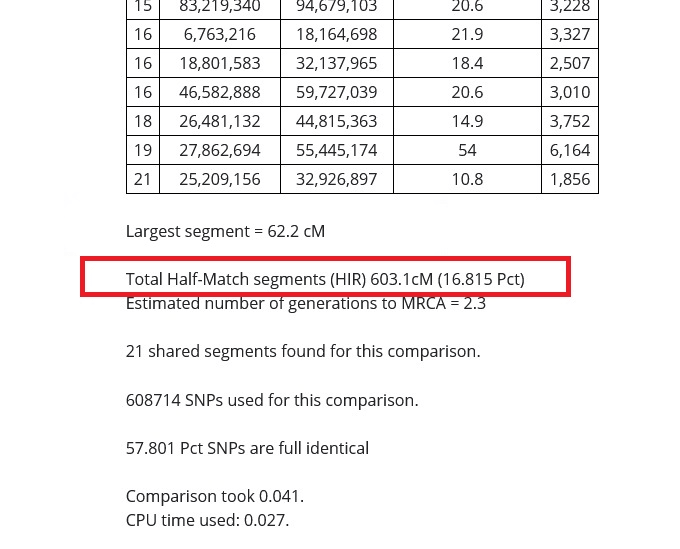
The reason that the total number of centimorgans (cMs) shared is important is because this information can help us estimate how closely related we are to the DNA match. The total of the half-match segments and the size of the longest or largest DNA segment shared are the most important details that we can use to learn about our connection to our match.
The total number of cMs shared is important because it gives you a clue as to how closely or distantly you are related. In general, the larger the number of cMs, the closer the relationship.
If you share lots of small segments with someone, however, it is possible that you are related in more than one way, especially if you know that you have endogamy in your family tree. It's a mind-bending thought, but it is possible to be 4th cousins once-removed AND 6th cousins, twice-removed with the same person, just on different lines.
DNA can really help you figure out things like this.
The tool is set to only pick up on segments that are larger than 7 cMs, and it will provide you the total number of segments that match this criteria. This is important because some segments smaller than 7 cMs are "coincidentally" identical to others, and other smaller segments are valid but too far back to reliably trace, even under the best of circumstances.
Finally, you might wonder what "HIR" stands for. These are half-identical regions in our DNA, which means that these segments are only identical on one copy of the chromosome.
We inherit one copy of each chromosome from both our mother and father. When we have a HIR (half identical region) shared with a match, it simply means that the match is only related to us on one side of our family.
In the case of my DNA match, my first-cousin once-removed, I know that he is related on my dad's side of the family. This is why we only have half-identical regions.
Is there such a thing as fully identical regions? Yes, for sure! We are most likely to share those with our full siblings, since there is a higher probability that some of the DNA segments that we share with them are identical on both copies because we are related on both sides of our family.
Number of matching segments
As shown in the image below, the number of matching segments that I share with my dad's first cousin is 18. I share many more segments with my sister and parents, however, and fewer with my more distant relatives.
The takeaway from this, as explained in the previous sections of this article is that the more segments that you share, the closer the relationship. This is especially true if the total amount of segments is high, and the size of the largest segment is high.
To summarize, in generall:
- Large number of matching segments, and a few large segments = close relationship
- Small number of matching segments, but at least one large segment = it is still possible to be a relatively close relationship
- Single small matching segment or a few small (2-3) matching segments = likely a distant relative
- Large number (5+) of matching small segments = possibly related in more than one way, perhaps distantly
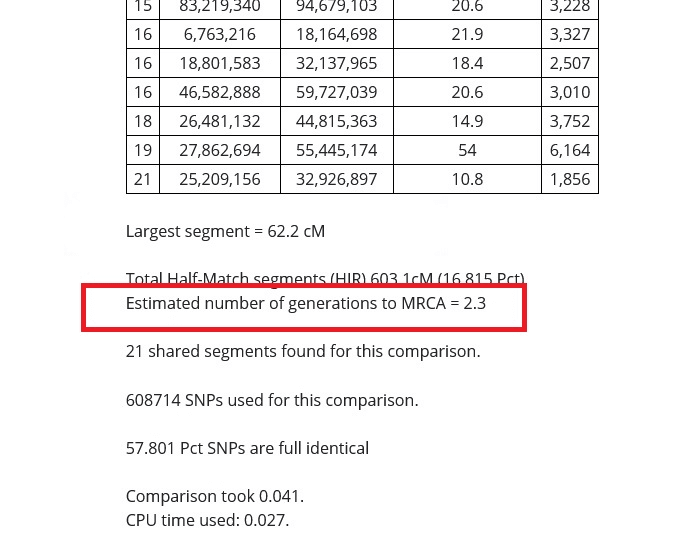
Estimated number of generations to MRCA
On Gedmatch results, we often see the estimated number of generations to the Most Recent Common Ancestor (MRCA). This is an estimated number of generations that we would need to go back in our family tree to find the most recent ancestor that we share in common with our DNA match.
The last number you see in the example that I shared is "Estimated number of generations to MRCA", and in my comparison with my cousin, it is 2.3 This is actually really very accurate in this particular case, since I know that my great-grandparents are his grandparents.
This tool is usually very accurate in estimating relationships when there are no "half" relationships involved and the relationships are close. Full siblings, parents, full cousins and second cousins, for example, will usually fall within the correct estimated number of generations on this tool.
The problem lies when the tool is trying to estimate generations between more distant cousins. It's possible for 4th cousins to share higher amounts of DNA than usual, for example, and in this case, the tool could throw you off and lead you to believe that you are actually more closely related than you are.
It's also very tempting to want to believe that the estimate is correct, too, since it's much easier to search through 8 great-grandparents searching for a common relative than trying to sift through 64 great-great-great-great grandparents, assuming that they are all known, which isn't always the case.
The MRCA estimate is useful, but the software is only basing its estimate on the number, size, and total of shared segments and making its best guess. When we are understanding the results, we need to take that into consideration.
SNPs Used For This Comparison
The number you see after "SNPs used for this comparison" tells us the number of SNPs, which stands for Single nucleotide polymorphisms, that were compared to determine the results. The higher the number, the higher the accuracy of the results.
Conclusion
I hope that this article helped shed some light on what you can learn from the Gedmatch One-to-One Autosomal DNA Comparison tool. If you have any questions about what you have seen in your own comparisons, I would love to hear from you in the comments.
Thank you so much for stopping by!


Robyn Telles
Monday 20th of May 2024
in this summary below please advise me
what number is considered a high centimorgan?
what number is considered a low centimorgan?
what number is considered large number of matching segments?
what number is considered small number of matching segments?
what number is considered a large segment?
what number is considered a small segment?
To summarize, in generall:
Large number of matching segments, and a few large segments = close relationship Small number of matching segments, but at least one large segment = it is still possible to be a relatively close relationship Single small matching segment or a few small (2-3) matching segments = likely a distant relative Large number (5+) of matching small segments = possibly related in more than one way, perhaps distantly
GC
Saturday 23rd of July 2022
Thank you for this explanation. I'm still partially confused or maybe more than partially. This is the 2nd closest match for me on gedmatch. Does this mean this is a 1st cousin once removed with 433.9cm? I think this person is related on one side and I'm guessing it must be my unknown bio father's family, yet I'm still confused!
Largest segment = 55.5 cM
Total Half-Match segments (HIR) 433.9cM (12.106 Pct) Estimated number of generations to MRCA = 2.5
16 shared segments found for this comparison.
154747 SNPs used for this comparison.
52.232 Pct SNPs are full identical
Mercedes
Sunday 24th of July 2022
Hi GC, Thank you for this question! This relative that you have found shares a lot of DNA with you, but they could be related in a few different ways. First cousin once-removed is certainly an option, but they could also be a half-first cousin (sharing only one grandparent with you), a half-sibling of your grandparent, or a couple other relationships. Do you know how old this person is? This could help eliminate some of the options. You might find this post helpful for narrowing down your search: https://whoareyoumadeof.com/blog/strategies-for-finding-biological-grandparent-using-dna/ I hope this helps, Mercedes
How to Understand the Gedmatch Are Your Parents Related Tool Results - Who are You Made Of?
Thursday 28th of January 2021
[…] have done DNA tests, you don’t need to use this Gedmatch tool. Instead, you can use the Gedmatch One-to-One tool to see if your parents share matching DNA […]