One of the unique challenges that you might encounter while researching your Mexican DNA matches is endogamy. What in the world is endogamy, and what does it have to do with Mexico and DNA matches? Endogamy is simply a fancy word that means intermarriage within a particular community or area. Basically, it's just the practice of marrying people within the town or village that a person grew up in. You usually don't marry people you don't know, of course, so throughout history, it has been a common practice - not just in Mexico, but all over the world.
I should note that this discussion in this article is specific to DNA matches and people with Mexican ancestry, but the concepts here are relevant and applicable to any population with endogamy, including other Native American groups, French Canadians, Colonial Americans, European Jews, and many others too numerous to list.
Endogamy and Native American DNA
If endogamy occurs over many generations, with little or no genetic input from distant, outside communities, most people within a particular area might end up being related to each other, sometimes in more than one way. In Mexico, and in many places in North and South America, just like in many other regions, endogamy occurred for the same reasons that it occurred in Europe, Africa or Asia. Additionally, the entire populations of North and South America, prior to the arrival of Europeans, were descended from a few small groups of people who migrated from Asia to North America over the Bering Strait.
With little to no outside genetic influence, and thousands of years of endogamy, this means that most people living in Mexico (if not all) were related to each other - albeit very, very distantly - in many different ways. Sometimes, people still have small DNA segments from these very distant ancestors. Sometimes, people still have many, many small segments from these very distant ancestors, and they might share these same very small, very numerous DNA segments with DNA matches.
How does endogamy affect DNA matches?
If you have been researching DNA, you might be familiar with the idea that you can estimate your relationship to a DNA match based on shared DNA. This is true, but the same rules don't apply to endogamous populations. And since the DNA testing companies don't necessarily know that your DNA was inherited from endogamous populations, this can mean that you share higher amounts of DNA (made up by these smaller, numerous segments inherited from many very distant ancestors) with your DNA matches, implying a closer genealogical relationship than you actually have.
Endgomay makes it more difficult to sort through matches and figure out which matches are good candidates for actually being able to find a connection. The image below is from my husband's DNA matches on Family Tree DNA. I'm definitely not picking on Family Tree DNA, however, since I have seen this exact issue on every single testing company. The example below is just an exceptionally excellent example of how endogamy can lead you to believe you are more closely related to a match than you actually are:
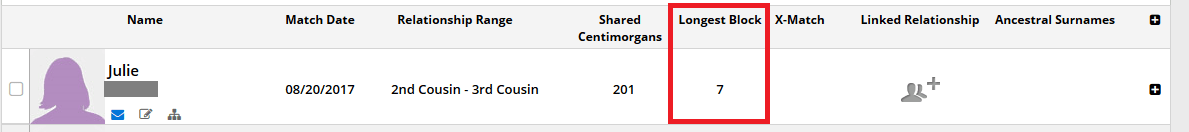
If you weren't careful, you could assume that my husband and Julie are relatively close cousins. Their relationship is estimated by the testing company to be a 2nd-3rd cousin relationship, since they share a total of 201 centimorgans (cMs) of DNA. If you look carefully, however, you will notice that I drew a red rectangle around the longest block of shared DNA and you can see that the longest DNA segment that they share is only 7 cMs!
A 7 cM segment implies a very, very distant relationship. While it's true that there is a small chance that it could be a match from having great-great-great grandparents in common, it is much more likely that Julie and my husband have a 9th great-grandparent in common. What's more interesting is that Julie and my husband share 194 additional centimorgans of additional DNA, not including the 7 cM segment. I took a look at the segments and all of them are much, much smaller. There is a good chance that Julie and my husband have 50 DNA segments in common!
There is a possibility that many of those segments are identical by chance (i.e. coincidentally identical, since the probability for this is increase when dealing with very small segments), but there is also a decent chance that Julie and my husband share a few dozen common ancestors many generations back in time. My husband and Julie are almost definitely related to each other in some way, or even in many ways, but their connection(s) is (are) so far back in time that the odds are their trees will never be complete or accurate enough to actually locate the common ancestor(s).
Endogamy and the number of DNA matches
One other very interesting aspect of endogamy is that it can make it appear that you have many more relatively close DNA matches than you actually do. Many people who have ancestry in endogamous populations report having thousands of 4th cousin matches or closer on their DNA match list. As you now know, many of these 4th cousin matches are not really 4th cousins, but they happen to share enough DNA to fall within the typical 4th cousin range, and thus, are categorized as such.
Some of these cousins will actually be 4th cousins, some closer, and some more distant. In order to really understand how an individual match is related, or to know whether it will be possible to do so, you'll have to take a close look at the exact amount of DNA that you share with a match, how many segments you share, and their family tree, if they have one.
How to correct for endogamy in DNA matches?
If endogamy makes all of your DNA matches appear to be more closely related than they actually are, is there any way that matches can still be a helpful tool in genealogical research? Absolutely! As I mentioned in the previous section, the size of the largest DNA segment can imply a distant relationship. This same bit information, the largest segment, can also be an indicator of a closer relationship.
DNA segments are broken up each generation. Let's take as an example a set of grandparents. The children of those grandparents will share very large segments (as high as 280+ centimorgans). When those children have children, their kids will inherit 50% of their DNA from each parent, meaning that some of those big segments that would match the grandparents will be passed down, but it smaller chunks. When the grandkids have children, the segments will become smaller yet. Each generation, these DNA segments will become smaller and smaller. This is exactly why we don't pay as much attention to the very small segments.
Since segments can break up pretty quickly, there is a good chance that segments that are larger than about 15-20 cMs are indicative - especially in endogamous populations - of an common ancestor that is within a genealogically verifiable time frame. With this said, the larger the segment, the better. In the image below, you can see another one of my husband's DNA matches:
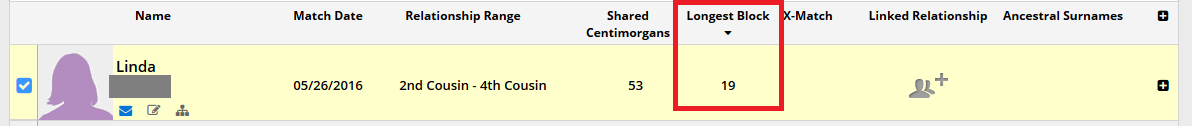
I highlighted the section that shows that Linda and my husband share 19 cMs of DNA on their longest segment. They share 53 centimorgans of total DNA, but all of those other segments are smaller than 5 cMs. There is a chance that some of those super small segments are legitimate, but they would be too far back to actually trace, so I won't pay attention to them. I would rather focus on the 19 cM segment because it could mean that Linda and my husband are 3rd-8th cousins. Depending on the exact relationship, I may or may not be able to find the ancestor that they share, but I actually have a decent chance of being able to do so.
You might be wondering, if you can find this same information about your matches on other sites. I know that I have been using Family Tree DNA as an example in this post, but you can access this information on most other websites where you have tested or uploaded your DNA.
Correcting for endogamy on My Heritage DNA
On My Heritage DNA you can sort your matches by Largest Segment:
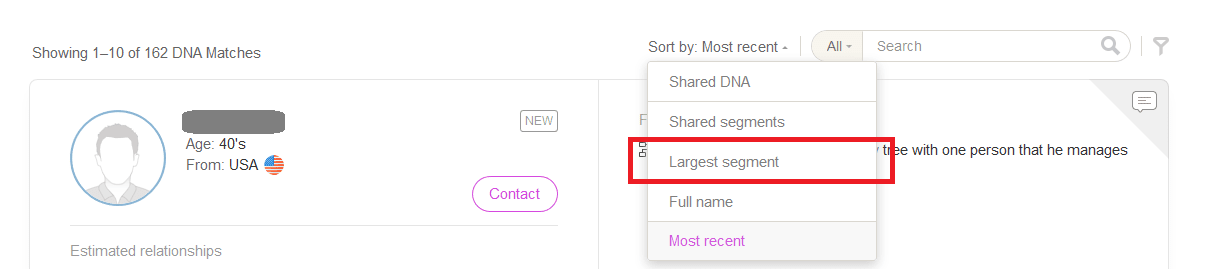
When I filter the results like this, I will now see DNA matches listed in order of largest shared segment to smallest segment, regardless of total amount of DNA shared, which is exactly what I want to see. I can now check the shared segment size, see which chromosome it is on, and see if the match has a family tree with surnames or locations that are familiar to me.
What about endogamy on Ancestry DNA?
It's a little more difficult to handle your DNA matches on Ancestry DNA and take engogamy into account, since we don't get to see the exact length of the shared DNA segments. We also can't filter or search through matches using criteria related to these more technical details. All hope is not lost, however, because we can still see how much DNA and the number of segments that are shared. Using this information, and the knowledge that Ancestry does not count segments smaller than about 6 cMs, in many cases we can infer this information.
(It's great that Ancestry ignores the very small segments - it take some of the work out for you - yay!)
For example, take a look at the match below:
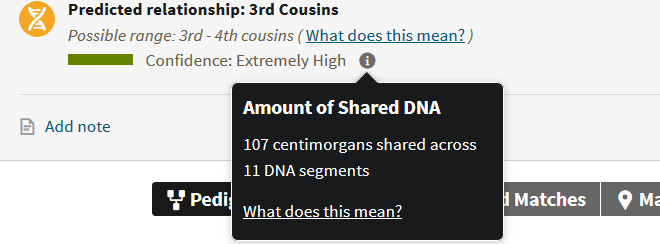
The total amount of shared DNA is 107 cMs, but it shared across 11 segments. If we divide them equally, that's less than 10 cMs per segment - meaning many distant ancestors. If we decide there may be one large one, and the rest are at the lowest threshold, this means that we could have one that is 47 cMs, and the rest are 6 cMs, meaning one relatively close ancestor (or set of ancestors) and several very distant connections. The reality is that the "truth" is probably somewhere between those two ideal scenarios.
The following match on Ancestry makes this exercise slightly easier:
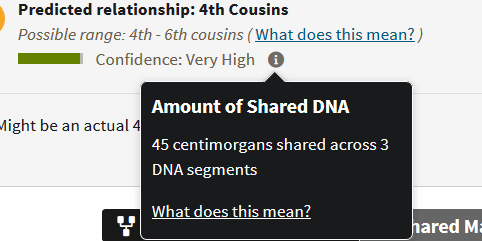
With this second match, you can see that he shares 45 segments across 3 segments. Even if you assume that it is equally divided, that is three segments of 15 cMs! Or if there are two segments of 6 cMs, that still leaves a nice segment of 33 cMs to work with, meaning that this particular match is a promising 3rd-5th cousin, even though they still might be related in more than one way (because of the multiple shared segments of a decent size).
Does this mean that I should automatically ignore small segments in my DNA matches?
You might be wondering if the presence of endogamy means that you should just automatically ignore distant cousins, or cousins with whom you share many small segments. Absolutely not! It's possible to share absolutely no DNA with a 3rd cousin, or to share only small segments (as a result of endogamy) with a third or fourth cousin, so I would definitely encourage you to examine each DNA match. You never know what you'll find, and what you'll learn.
Now that you understand why you share so many DNA segments with these cousins, you will not be distracted by the higher than usual amounts of shared DNA and will be able to look for more reliable evidence of a genealogically traceable connection.
Conclusion
I hope that this post has helped you understand a little bit about the unique challenge that endogamy presents for people with extensive Mexican ancestry. If you have any questions, comments, or concerns about something that you have read here, please feel encouraged to leave a comment below. I hope to hear from you.
Thanks for stopping by!



How to Know if You Have Endogamy in Your DNA Matches - Who are You Made Of?
Thursday 12th of November 2020
[…] example, take a peek at the image below. It’s a screenshot of my husband’s DNA match list on Family Tree […]